Segmentation model
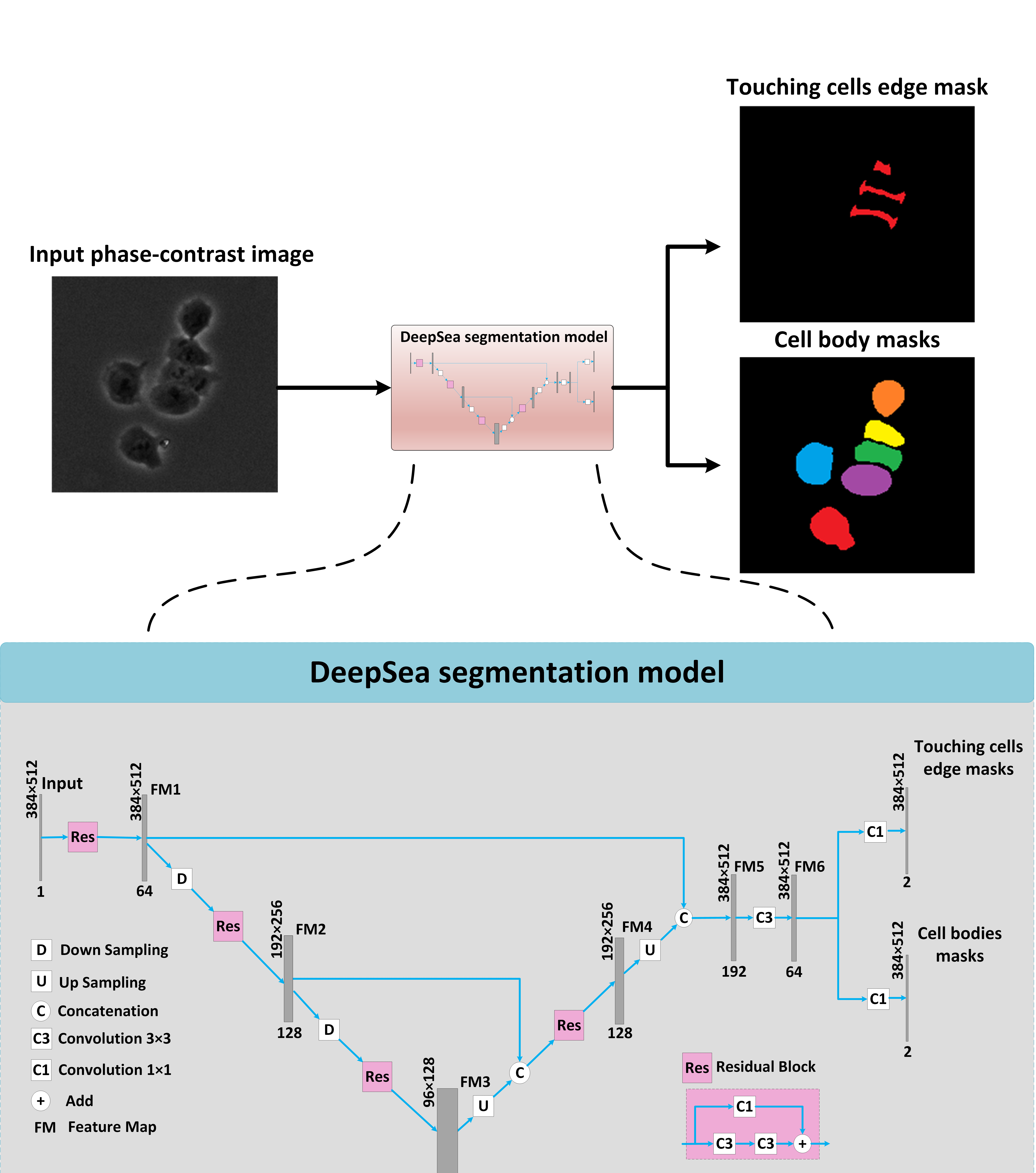
In the instance segmentation task, we proposed and built a 2D deep learning-based model called DeepSea (Figure 8). Our model employs residual blocks aiming to 1) increase the depth of the network resulting in fewer extra parameters instead of widening the network, 2) accelerate the speed of training of the deep networks, 3) reduce the effect of the vanishing Gradient Problem, 4) To potentially obtain higher accuracy in network performance as observed in Residual Networks. Our DeepSea model involves only 1.9 million parameters indicating that it is considerably smaller than typical instance segmentation models such as UNET, PSPNET, SEGNET, and so forth. We also took advantage of batch normalization and dropout techniques to improve the model’s speed, performance, and stability. The auxiliary touching cell edge representations (highlighting the edge area between touched cells) and the auxiliary training loss value we involved lead the learning algorithm to spend more computational budget and time on touching cell edges and better learn them during the training process.
Tracking model
Designing a cell tracking model aims to track cell bodies from one frame to the next and also detect cell divisions (mitosis). The cell tracking process helps create cell lineage structure over time. We used an architecture baseline similar to the DeepSea segmentation model but with multiple images, two inputs, and one output. The first input is the target cell image at the previous time point and the second input is the segmented cell image at the current time point, and the output is the binary cell body mask of the target cell at the current time point. This model extracts the features of the single target cell in the previous frame to localize and detect it among the segmented cells in the current frame. To increase the accuracy of the tracking model and help it get rid of extra non-useful background information, we make it search only a small square area (with the size of 6 times the target body size) around the target cell’s previous location in the current frame. Since cells move slowly through space, the cell’s previous location will present a good guess of where the model should expect to find it in the current frame. The number of the model parameter is only 2.1 million, while the other most used tracking models, such as ROLO, DeepSort, TrackRCNN, use more than 20 million parameters, confirming that we have an efficient model in the tracking process as well.

